Feature Importance on MNIST with feature counts¶
Select a subset of 3’s and 5’s from MNIST¶
Here we train a forest using RerF with the RandMatImagePatch
option with patch.min = 1 and patch.max = 5.
## Get a random subsample, 100 each of 3's and 5's
threes <- which(mnist$Ytrain %in% 3)
fives <- which(mnist$Ytrain %in% 5)
numsub <- c(threes, fives)
Ytrain <- mnist$Ytrain[numsub]
Xtrain <- mnist$Xtrain[numsub,]
Ytest <- mnist$Ytest[mnist$Ytest %in% c(3,5)]
Xtest <- mnist$Xtest[mnist$Ytest %in% c(3,5),]
# p is number of dimensions, d is the number of random features to evaluate, iw is image width, ih is image height, patch.min is min width of square patch to sample pixels from, and patch.max is the max width of square patch
p <- ncol(Xtrain)
d <- ceiling(sqrt(p))
iw <- sqrt(p)
ih <- iw
patch.min <- 1L
patch.max <- 5L
forestS <- RerF(Xtrain, Ytrain, num.cores = numCores, FUN = RandMatImagePatch,
paramList = list(p = p, d = d, iw = iw, ih = ih,
pwMin = patch.min, pwMax = patch.max), max.depth = 8)
predS <- Predict(Xtest, forestS, num.cores = numCores)
(mnist.error.rate <- mean(predS != Ytest))
## [1] 0.01892744
forestRerF <- RerF(Xtrain, Ytrain, num.cores = numCores, FUN = RandMatBinary,
paramList = list(p = p, d = d), max.depth = 8)
predRerF <- Predict(Xtest, forestRerF, num.cores = numCores)
(mnist.error.rate <- mean(predRerF != Ytest))
## [1] 0.02208202
forestRF <- RerF(Xtrain, Ytrain, num.cores = numCores, FUN = RandMatRF,
paramList = list(p = p, d = d), max.depth = 8)
predRF <- Predict(Xtest, forestRF, num.cores = numCores)
(mnist.error.rate <- mean(predRF != Ytest))
## [1] 0.02050473
system.time({
FeatImpS <- FeatureImportance(forestS, num.cores = numCores, type = "C")
})
## Message: Computing feature importance as counts of unique feature combinations.
## user system elapsed
## 1229.062 6.560 314.241
ni <- matrix(0, length(FeatImpS$features), ncol(Xtrain))
for(i in 1:length(FeatImpS$features)){
ni[i, FeatImpS$features[[i]]] <- FeatImpS$imp[i]
}
NN <- matrix(apply(ni, 2, sum) / nrow(ni), 28, 28, byrow = FALSE)
saveRDS(NN, file = "NNsrerf.rds")
system.time({
FeatImpRerF <- FeatureImportance(forestRerF, num.cores = numCores, type = "C")
})
## Message: Computing feature importance as counts of unique feature combinations.
## user system elapsed
## 1750.585 11.082 444.860
nir <- matrix(0, length(FeatImpRerF$features), ncol(Xtrain))
for(i in 1:length(FeatImpRerF$features)){
nir[i, FeatImpRerF$features[[i]]] <- FeatImpRerF$imp[i]
}
NNr <- matrix(apply(nir, 2, sum) / nrow(nir), 28, 28, byrow = FALSE)
saveRDS(NNr, file = "NNrerf.rds")
system.time({
FeatImpRF <- FeatureImportance(forestRF, num.cores = numCores, type = "C")
})
## Message: Computing feature importance as counts of unique feature combinations.
## user system elapsed
## 14.579 0.323 5.549
nirf <- matrix(0, length(FeatImpRF$features), ncol(Xtrain))
for(i in 1:length(FeatImpRF$features)){
nirf[i, FeatImpRF$features[[i]]] <- FeatImpRF$imp[i]
}
NNrf <- matrix(apply(nirf, 2, sum) / nrow(nirf), 28, 28, byrow = FALSE)
saveRDS(NNrf, file = "NNrf.rds")
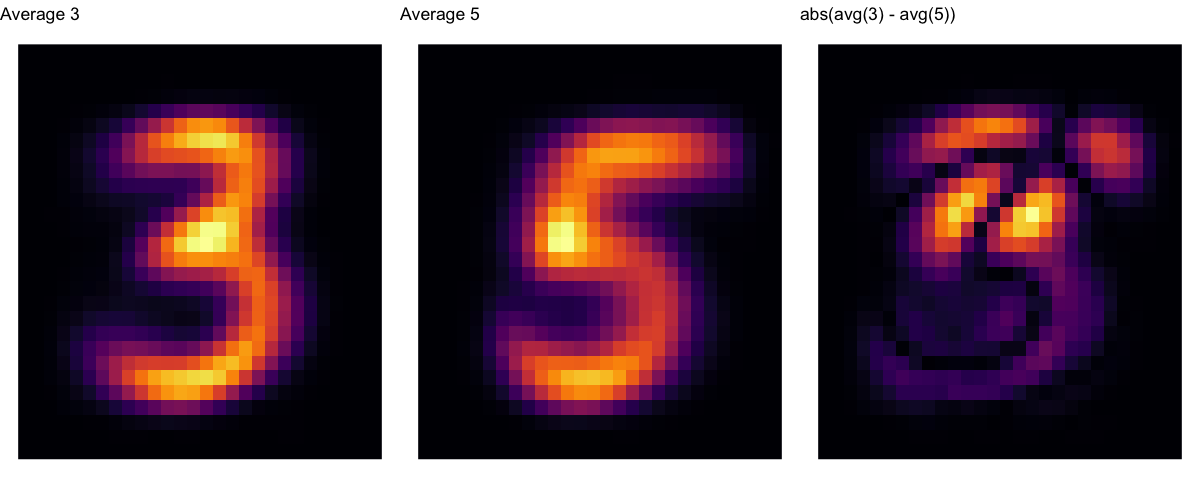
plot the average 3 and 5 from the training set¶
the3s <- Xtrain[Ytrain == 3, ]
the5s <- Xtrain[Ytrain == 5, ]
sum3 <- matrix(apply(the3s, 2, sum) / sum(Ytrain == 3), 28, 28, byrow = FALSE)
sum5 <- matrix(apply(the5s, 2, sum) / sum(Ytrain == 5), 28, 28, byrow = FALSE)
g <- expand.grid(x = 1:28, y = 1:28)
gg <- rbind(g, g)
nn <- as.vector(t(NN[28:1, ]))
nnrerf <- as.vector(t(NNr[28:1, ]))
nnrf <- as.vector(t(NNrf[28:1, ]))
s3 <- as.vector(t(sum3[28:1,]))
s5 <- as.vector(t(sum5[28:1,]))
s3m5 <- abs(s3 - s5)
Z <- data.frame(g, weight = c(nn, nnrerf, nnrf, s3, s5, s3m5), Alg = rep(c("MF", "Sporf", "RF", "Average 3", "Average 5", "x3m5"), each = length(nn)))
sc0 <- scale_fill_gradientn(colours = viridis(255))
sc1 <- scale_fill_gradientn(colours = inferno(255))
a1 <- ggplot(data = Z[ Z$Alg == "Average 3", ], aes(x = x, y = y, fill = weight)) + geom_raster() + theme_void() + guides(fill = FALSE) + sc1 + ggtitle("Average 3")
a2 <- ggplot(data = Z[ Z$Alg == "Average 5", ], aes(x = x, y = y, fill = weight)) + geom_raster() + theme_void() + guides(fill = FALSE) + sc1 + ggtitle("Average 5")
a3 <- ggplot(data = Z[ Z$Alg == "x3m5", ], aes(x = x, y = y, fill = weight)) + geom_raster() + theme_void() + guides(fill = FALSE) + sc1 + ggtitle("abs(avg(3) - avg(5))")
grid.arrange(a1, a2, a3, ncol=3)
#ggslackr(grid.arrange(a1, a2, a3, ncol=3), channels="#manifold-forest")
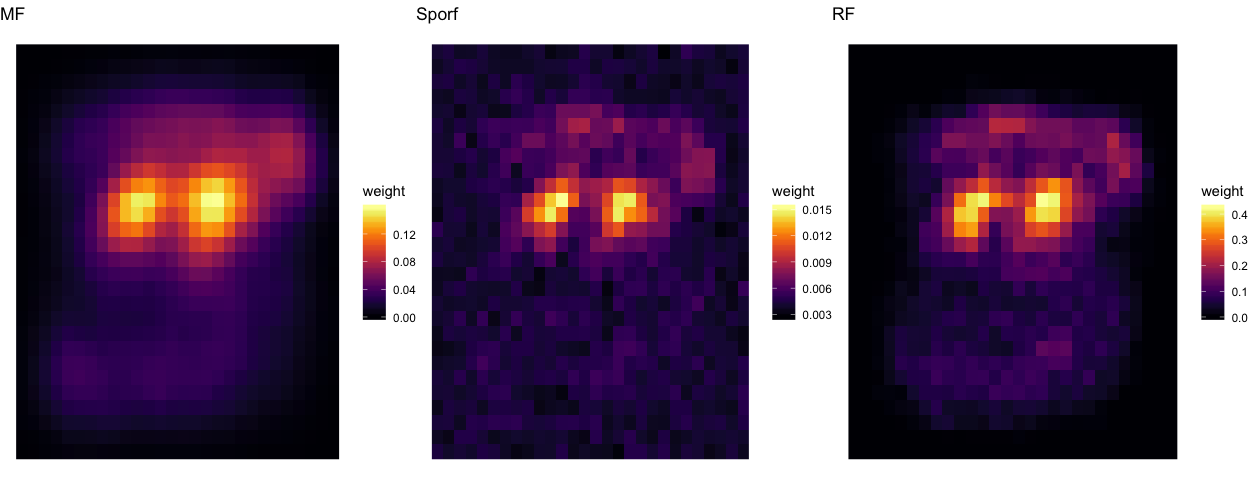
Feature heatmap¶
These are the features that S-RerF and RerF used, plotted as averaged heatmaps.
p1 <- ggplot(data = Z[ Z$Alg == "MF", ], aes(x = x, y = y, fill = weight)) + geom_raster() + theme_void() + sc1 + ggtitle("MF")
p2 <- ggplot(data = Z[ Z$Alg == "Sporf", ], aes(x = x, y = y, fill = weight)) + geom_raster() + theme_void() + sc1 + ggtitle("Sporf")
p3 <- ggplot(data = Z[ Z$Alg == "RF", ], aes(x = x, y = y, fill = weight)) + geom_raster() + theme_void() + sc1 + ggtitle("RF")
grid.arrange(p1, p2, p3, ncol=3)